Decoding Wild Rice Genomes: A New Era for Climate-Resilient Agriculture
Asian rice, domesticated around 10,000 years ago, is a dietary staple for more than one third of the global population. Yet, intensive breeding for improved yield has reduced its genetic diversity – making modern varieties increasingly vulnerable to stressors like heat, drought, and salinity. In contrast, wild rice relatives have adapted to a wide range of environmental conditions providing untapped genetic material for rice crop improvement.
A landmark study, published in Nature Genetics on April 28, 2025, traced the evolutionary history of wild rice species, classified in the genus Oryza, defining their phylogenetic relationships and deciphering how their genomes have been shaped by 15 million years of evolutionary history. The research was led by Prof. Rod Wing, Director of the Arizona Genomics Institute (AGI), and his postdoctoral research associate, Alice Fornasiero, of the King Abdullah University of Science and Technology (KAUST) in Saudi Arabia, in collaboration with Wageningen University & Research in the Netherlands.
Thanks to the sequencing of both genomic DNA and RNA of 11 wild rice species (i.e. two diploids and nine tetraploids) using the PacBio Sequel II and IIe instruments, the team could successfully generate ultra-high quality genome assemblies and use accurate full-length transcripts for gene annotation. The analysis of this valuable and publicly available genomic resource allowed the construction of the Oryza pangenome – i.e. the genomic representation of the entire genus – and the description of a “core” portion remarkably stable and a “dispensable” portion mainly composed of transposable elements. Due to their ability to move and reshuffle the genome, these elements has played a crucial role in shaping the genetic diversity in these species.
The analysis of gene expression in Oryza coarctata, a species adapted to saline conditions of coastal regions, showed an unusual balance between its two subgenomes, with both contributing equally to gene activity. This equilibrium may help explain the resistance of O. coarctata to salty environments.
These findings build a genetic foundation that could help scientists breed more resilient rice varieties, or even domesticate wild species through approaches like neodomestication to generate climate-ready crops for a more sustainable agriculture.
Use of Sage technology at the Arizona Genomics Institute’s Service Center:
For the wild rice genome project described above, the genomes were sequenced in both CLR and CCS/HiFi modes. The WGS libraries were size selected with the BluePippin, using 0.75% agarose cassettes with marker U1 (for CLR libraries) or marker S1 (for CCS/HiFi libraries ).
For current projects at AGI, the team has switched from the BluePippin to the PippinHT for its size selection needs. AGI has been an early access test site for the PippinHT Range+T program mode (for improving accuracy of HMW size selection) and the soon-to-be-released PippinHT 0.75% agarose cassette internal standards (which increase sample capacity).
“Range+T “ for Tight Sizing of HMW Libraries
Range+T Size Selection
Note: Range + T is available for all PippinHT instruments. However, only BluePippins with serial numbers 2700 and above will run the method.
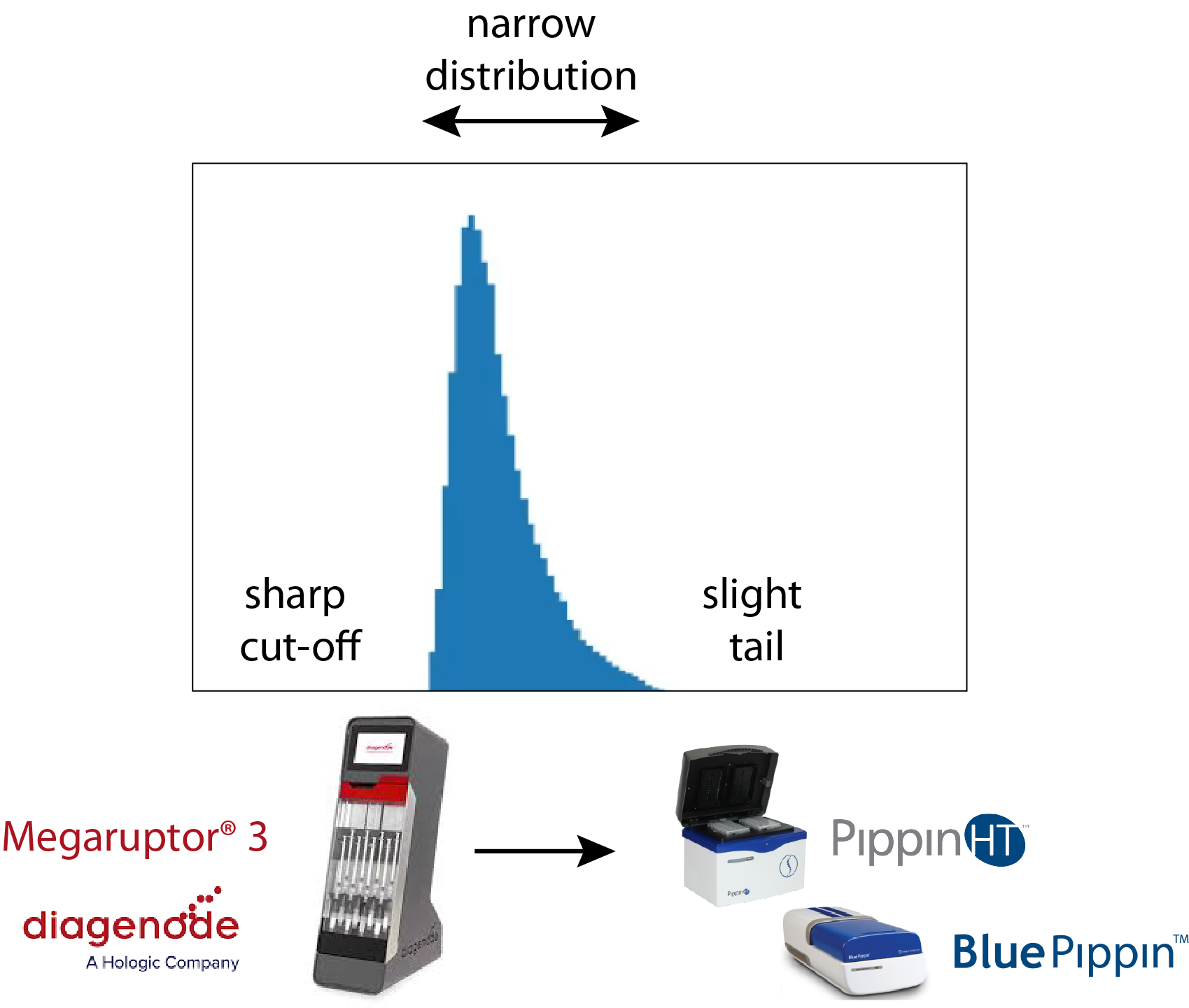
Sage’s DNA size selection technology has been used to help improve PacBio sequencing, particularly for mammalian and complex genome work since the introduction of the BluePippin (2012). Over the last few years, PacBio®’s Hi-Fi circular consensus sequencing has been the method that has set the gold standard for whole genome sequencing. In terms of library size, this technique benefits greatly from a sharp size cut-off at the LMW end of the size distribution (~10kb) and a slight tail at the HMW end. It also benefits from a narrow fragment size distribution around 17-18kb. A common way to accomplish this is to shear DNA with Diagenode’s Megaruptor®3 device, followed by size selection on a PippinHT or BluePippin.
To get the desired fragment distribution at these size ranges is not trivial. In Sage gel cassettes, the total input load – and the tight fragment distribution profile of Megaruptor 3 sheared samples – affects the mobility of the DNA in a manner that cannot be easily accounted for with our standard “Range” programming method. Essentially, the start of collection of DNA is accurate and straightforward, while the timing for the end of collection can be affected by the physical nature of the sheared DNA sample. To this end, we developed a new size selection method called “Range+T”. With this method, the base pair start of collection is entered in the software and the end of collection is determined by entering a time for elution. Through a little trial and error, a time setting can be determined to overcome calibration uncertainty for the end of collection. We conducted an early access program with the Copetti lab at the Arizona Genomics Institute, resulting in increased PacBio HiFi read lengths, that you can read about in this application note.
A Collaboration with Diagenode
We decided to do a deep dive into Range+T to get a better handle the method, and to develop best practices for using Diagenode Megaruptor 3 and PippinHT or BluePippin. Working with Diagenode, we were able to process many DNA samples and experiment with multiple settings. We outlined our findings in a new application note, Best practices: Sage Science size selection with Diagenode Megaruptor shearing for long-read sequencing library preparation . We encourage you to take a look if you are interested in working with these size ranges. We found that careful coordination of Megaruptor shearing and Sage size-selection conditions are critical:
1. Megaruptor shearing conditions should ideally be chosen to provide a sheared DNA input with an average size at, or slightly above, the desired library size.
2. For accurate LMW cut-offs, start of collection DNA size on PippinHT or BluePippin should be less than or equal to the peak size (mode) of the sheared DNA input.
3. The upper boundary of the size distribution will be determined by the Megaruptor shear conditions if long Sage Range+T elution times are used (i.e., 30 minutes or longer).
4. Sage Range+T programming can provide significantly narrower size distributions by shortening the Range+T elution time below 30 minutes.
For size selection, provided the same Megarupter shear conditions (and input amount) are used, we recommend starting with Time values at 5 minute intervals (ie. 5,10,15,20 minutes) to determine the best distribution conditions. It is very important to enter a starting base pair value that is before the mode of the Megaruptor distribution – so QC your samples!
Range+T is available now for both PippinHT and BluePippin. It requires a software upgrade (v.6.41 and v1.14, respectively, available for download on sagescience/support). We have also packaged cassette kits specifically for this purpose; HRT7510 and BRT7510. Starting Range values can be between 9-30kb.
Cut the Big Stuff: >50kb High Pass DNA Size Selection on the HLS2
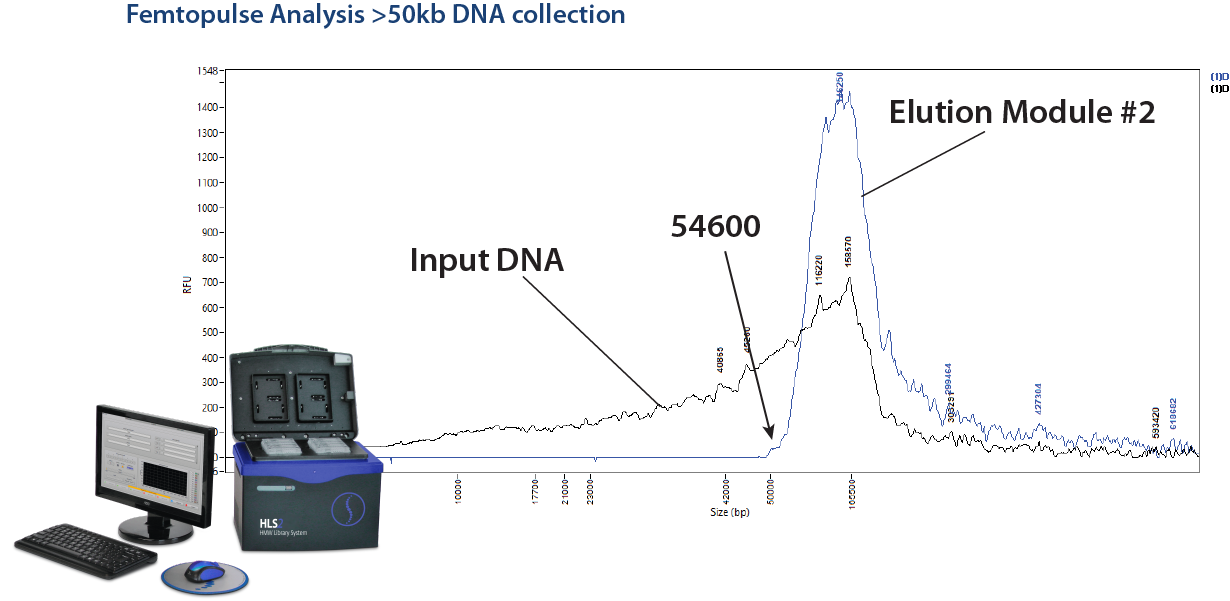
We have been getting quite a few inquiries of late from researchers who would like to size select DNA fragments above 50kb. This may be due in no small part to our friends at Grandomics, China, who have been citing this capability in a number of interesting collaborative studies (see the reference links below). In short, this is something that cannot be accomplished on a BluePippin, and Grandomics has been using the HLS2 platform to accomplish this feat. Here at Sage, we have compiled a cassette kit (HSS-0012 or HSS-0004) specifically for this, and for DNA size selection in general. In fact, with direct current electrophoresis, the HL2 can size select DNA in ranges between 1-20kb, and with pulsed-field protocols it can size select Ultra-HMW DNA into the hundreds of KB and up to 2MB.
HLS2 Size selection on the HLS2 is outlined in this application note: Ultra-HMW DNA Size Selection with the HLS2 instrument and a >50kb Hi-Pass protocol. A quick note to Oxford Nanopore users: this method uses a starting input of 5 ug of DNA and yields about 1-1.5 ug of DNA, while some Oxford Nanopore input requirements may be quite a bit higher.
References:
Huang, Z., et al. 2023. Evolutionary analysis of a complete chicken genome. PNAS 12 (8) e2216641120.
https://doi.org/10.1073/pnas.2216641120
Morita, S., et al. 2023. The draft genome sequence of the Japanese rhinoceros beetle Trypoxylus dichotomus septentrion towards an understanding of horn formation Sci Rep 13, 8735 (2023).
https://doi.org/10.1038/s41598-023-35246-w
Xing Guo, et al. 2023. The genome of Acorus deciphers insights into early monocot evolution. Nat Commun 14, 3662 (2023).
https://doi.org/10.1038/s41467-023-38836-4
Kun Li, et al. 2023. Genetic Diagnosis of Facioscapulohumeral Muscular Dystrophy Type 1 Using Rare Variant Linkage
Analysis and Long Read Genome Sequencing. medRxiv preprint.
https://doi.org/10.1101/2023.06.05.23290975
Q Wang, et. al. 2023. Draft genome of the oriental garden lizard (Calotes versicolor). Front. Genet., 20 February 2023
Sec. Livestock Genomics Volume 14 – 2023
https://doi.org/10.3389/fgene.2023.1091544
Fengjiao Ma, et al. 2023. Gap-free genome assembly of anadromous Coilia nasus. Sci Data 10, 360 (2023).
https://doi.org/10.1038/s41597-023-02278-w
AGBT23 Roundup!
Well, we have another fabulous AGBT behind us that we were happy to attend. Our take home messages? A. Spatial transcriptomics is the exciting new frontier. B. True human WGS has been accomplished and is moving into population-scale work. And C. There are a lot of new sequencing technologies being bandied about.
Though we weren’t presenting or introducing, Sage Science did have a few key mentions. The Broad Institute described their efforts to automate a PacBio HiFi whole genomes sequencing pipeline for the NIH’s All of Us program. In their poster, you can view it here, they go into quite a bit of detail why gel size selection (via the PippinHT) is the preferable method with HiFi library construction.
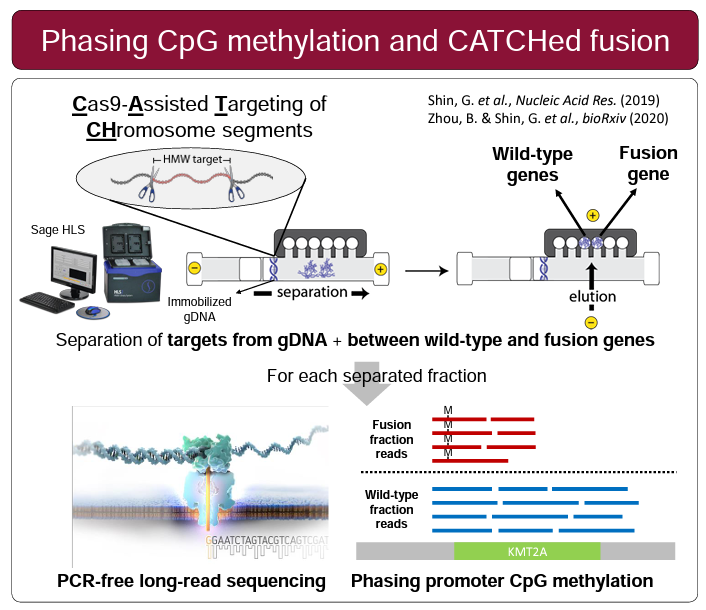
HLS-CATCH, our method for enriching high molecular weight targets made an appearance in a few posters. Universal Sequencing, our frequent collaborators and partners (we also sell their TELL-seq kits) presented a nice demonstration of their linked-read technology. They show haplotype phasing data for larger targets (200kb with HLS-CATCH) and smaller targets (4-20kb) using PCR and andCas9 Exo/Pulldown method (a modified CaBagE protocol).
Three posters were presented that featured HLS-CATCH and Oxford Nanopore sequencing. Researchers from the Ji lab at Stanford University presented a very compelling poster on KMT2A translocations, a common chromosomal abnormality in leukemias. Using HLS-CATCH (they use the term CLTR-Seq) and nanopore sequencing they identify rearrangement structure as well as the methylation landscape. Stanford researchers from the Ji and Urban labs presented a poster on a study in which HLS-CATCH is used to sequence segmental duplications, also by nanopore sequencing, associated with the neuropsychiatric 22q11.2 deletion region. Finally, CATCH was used in a study presented by Gavin Arno from the University College London and collaborators that is focused on inherited retinal disease and complex rearrangements associated with the OPN1LW/OPN1MW gene array. The authors, like the Stanford researchers, conclude that CATCH-nanopore sequencing is effective unraveling genomic mysteries that are “intractable to NGS”.
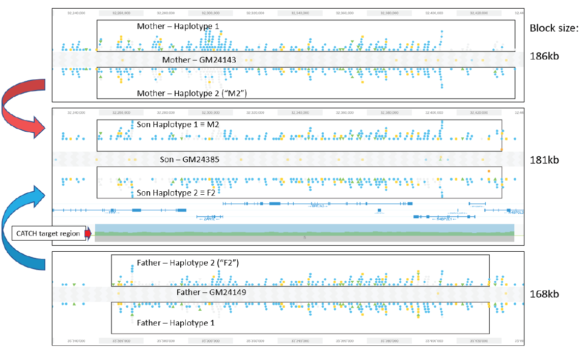
HLS-CATCH and TELL-Seq: Haplotype Phasing of a 187kb HMW Gene Targets from a Trio
We here at Sage Science are excited to have joined forces with Universal Sequencing Technology to promote their TELL-Seq barcode linked-read technology. One of the reasons for our excitement is that TELL-Seq, and the transposase-based barcoding method benefits greatly from high molecular weight DNA – one of Sage’s areas of expertise. Our own HLS-CATCH method is one area that is a particularly nice fit with TELL-Seq particularly given the relatively small amount of DNA that HMW target purification yields. The easy library workflow and ready access to Illumina sequencing is also a big plus.
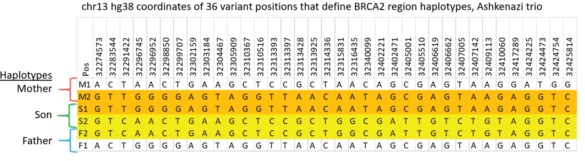
Validating this proposition was simple and straightforward. We performed HLS-CATCH using cultured cells from the Genome in a Bottle Ashkenazi trio sample set using a Cas9 guides that we designed to purify a 187kb target containing the BRCA2 locus. TELL-seq libraries were prepared from the targets from each individual, sequenced and run through 10X Genomic’s Long Ranger analysis package and then visualized the data on the 10X Loupe browser. The phase blocks sizes were 186 kb (mother), 181kb ( son), and 168Kb ( father), and only a small-genome scale analysis was required. Download our whitepaper here.
The variants in the 6 haplotypes compared favorably to the Genome in a Bottle high quality sequences.
Clearly CATCH and TELL-seq can be an economical alternative for obtaining long range genomic data for gene targets. The value of this technique was previously demonstrated in a Stanford University paper (Shin, GW, et al. 2019) where, among other analyses, the 4Mb MHC histocompatibility complex was phased using the 10X Genomics linked read method. The simplicity of TELL-Seq is great news vis a vis the effort required for long-read sequencing libraries. However, strides are being made there as well. Two recent papers have demonstrated great results using HLS-CATCH with PacBio sequencing (Walsh, T. et al. 2020) and Oxford Nanopore (Zhou, B, et al. 2020).