Cell Paper Reports Massive Genome Study for Lung Cancer
A paper that came out last month in Cell describes a massive effort to generate the exome and/or genome sequences of 183 tumor/normal pairs for people with a specific type of lung cancer. Authors hailed from Harvard University, New York University, the University of Cologne, MIT, and many other institutions.
Through this major collaboration to examine lung adenocarcinoma, for which affected patients have a five-year survival rate of about 15 percent, scientists “identified novel mutated genes with statistical evidence of selection and that likely contribute to pathogenesis,” they write in the paper. The genes implicated in lung adenocarcinoma from this study corroborate earlier links between this disease and many of the genes. By grouping variants together, the study also found signatures that correlate with a patient’s history of smoking as well as alterations of genes linked to the cancer.
In addition to the biological advance in understanding this disease, the authors suggest that this may have relevance downstream as well. “The candidate genes identified in this study are attractive targets for biological characterization and therapeutic targeting of lung adenocarcinoma,” they write.
We were proud to see that this work included using our Pippin Prep platform for size selection. It’s wonderful to have our technology contributing to a study of this magnitude, and one that could ultimately help prevent some of the 500,000 deaths that occur globally each year due to lung adenocarcinoma.
Link to: Mapping the Hallmarks of Lung Adenocarcinoma with Massively Parallel Sequencing. Cell 150, 1107–1120, September 14, 2012.
What’s in a Name?
We frequently get asked about the name “Pippin”, and why we’ve chosen it for our products. We thought it would be fun to see if the moniker has been also adapted in other fields of use — this is what we found:
Peregrin “Pippin” Took: Born in the Third Age 2990 (or 1390 in Shire Reckoning) he went on to become a Knight of Gondor. Way more than you need to know about him can be found at this link.
Steve Peregrin “Pippin” Took: This Hobbit-loving hippie was a founding member (with Marc Bolan) of Tyrannosaurus Rex, which subsequently morphed into the British glam-band T. Rex. Also a member of the cult band Pink Fairies, his drug-addled life was cut short at age 31 after allegedly inhaling a cocktail cherry. You can check out some of his songs here.
Blue Pippin guitar: A solid-body electric guitar from Specimen Products, a Chicago-based instrument maker founded by sculptor Ian Schneller. The Blue Pippin features an attractive deep-blue finish, and they also make a very cool-looking axe named the Pale Blue Pippin. Seems like a great opportunity for co-promotion — any hard-rocking geneticists out there?
Zippin Pippin: Built in 1912 in Memphis, this is one of the oldest wooden roller coasters still in operation and is reported to have been Elvis Presley’s favorite amusement ride. The Zippin Pippin was salvaged and successfully relocated to Green Bay, WI, in 2010. This effort undoubtedly restored some karmic balance to an occasionally cruel world.
The Zippin Pippin before being given a second life.
Apple Bandai Pippin: Launched in 1996, the Bonzai Pippin was Apple’s foray into the videogame market. The product, a venture between Apple and toy-manufacturer Bandai, was supposed to be an inexpensive console for gaming and networking. Instead, it was a total flop. (Apparently this did not forever taint the company as a failure.) Apple stock was trading at about $22/share in 1996, in case you were wondering.
Cincinnati Pippins: In 1912, the Cincinnati Pippins were a baseball team that belonged to a league that survived for only one month. The Pippins won 12 games and lost 10, placing them ahead of the Chicago Green Sox.
Pippin the musical: Written by Stephen Schwartz, originally directed by Bob Fosse, and Ben Vereen won a Tony award. The piece is based on the eldest son of Charlemagne (circa 767-811), Pippin the Hunchback, without the hunchback or any historical basis — but with great tunes.
Then there’s Pippin the Invincible, the pug. A loyal companion of our esteemed CSO, Dr.Chris Boles. Other than the fact that Chris developed the Pippin Prep, there is little connection, other than coincidence, between these two Pippins…
If you come across any other neat uses of “Pippin,” let us know!
Scientists Use Pippin in Sequencing Effort for Seven Cholera Isolates
A paper published this month in BMC Genomics discusses findings from a collaborative project to sequence cholera samples linked to the 2010 outbreak in Haiti — and we’re pleased to report that the Pippin platform from Sage Science was used in the work.
Lead author Rachel Sealfon and senior author Pardis Sabeti, along with a team of scientists hailing from MIT, Harvard, the Broad Institute, and other organizations, sequenced seven isolates of Vibrio cholera, including four collected recently in Haiti and the Dominican Republic.
The paper, entitled “High depth, whole-genome sequencing of cholera isolates from Haiti and the Dominican Republic,” concludes that the outbreak strain found in Haiti and the isolate found in the Dominican Republic most closely match strains found in South Asia, confirming publications released last year that suggested the epidemic came from a South Asian strain of cholera brought to Haiti through human activity.
The scientists also note that part of their goal was to assess coverage needed for sequencing, particularly for this type of effort — that is, sequencing closely related isolates and identifying the variants distinguishing them. “We found that 50x coverage is sufficient to construct a whole-genome assembly and to accurately call most variants from 100 base pair paired-end sequencing reads,” the authors write.
Check out the paper and how the authors used Pippin here [PDF].
BluePippin “Mid-Range” Size Selection: 18-27 kb
We’re pleased to be shipping our Mid-Range size selection cassette (BMF7510) for the BluePippin. While our Low-Range cassette (BLF7510, 1kb-10kb) works for many mate-pair library construction protocols, we’ve been hearing from our customers that there is interest in creating larger circularized molecules. This cassette, which continues to extend the range and flexibility of the BluePippin product line, should serve as a useful tool for large-fragment preps and help produce more diverse and meaningful data.
(Some internally generated validation data can be found on this pdf.)
In this blog, we’d like to outline some characteristics on the product to help users develop appropriate methods.
CVs and target ranges
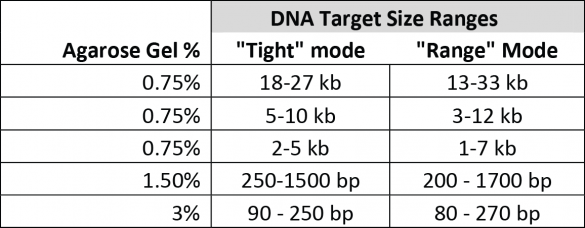
We define all target ranges in terms of what a user can accurately collect by entering a value in “tight” mode using our software. We also have a specification, “Minimum Size Distribution as Expressed by CV,” that describes the minimum distribution of fragments that may be collected given the capabilities of the system and the resolution of the gel. For instance, as a rule of thumb, a CV of 8% (our spec for smaller targets) will yield a range of fragments that are at most + 16% of the median fragment. However, the mid-range targets collect at a comparatively larger distribution minimum (20% CV), which limits the effective range of accurate “tight” cuts. This chart compares the accurately selectable “tight mode” ranges to the “range mode” ranges for the gel cassette calibrations available for the BluePippin at this time:
Sample Load Dependence
To calibrate the mid-range cassettes, we use a 5 ug sample load of sheared E. coli genomic DNA. Higher sample loads (up to 10ug) will usually run somewhat slower, relative to the marker DNA, and for this reason, you will need to program higher bp target or range values in the protocol to collect a desired size fraction. In our tests, when using a 10 ug input load, bp target values should be increased by 10-15% to compensate for the changes in mobility caused by the increased load.
Another factor to consider is the size distribution of the input DNA. For calibration and testing, we use an E. coli genomic DNA sample that has a very broad, almost flat size distribution. Using such samples simplifies our calibration process, which involves accurately sizing fractions sliced from the flat input samples. However, most input samples for mate-pair libraries are generated by methods that produce much narrower size distribution (DigiLab Hydroshear, for instance). Such samples will therefore show a different mobility dependence on input load than our E. coli genomic DNA test samples. For this reason, customers should plan on conducting some pilot experiments on non-valuable samples to investigate the mobility-load relationship produced by their specific library protocol.
Bottom line on 0.75% Mid-range cassette definition
- BluePippin 0.75% mid-range cassettes facilitate size fractionations of DNA samples out to 30kb.
- The CVs of tight size selections will be wider than those obtained from other BluePippin and Pippin Prep cassettes (mid-range CVs around 20%).
- The mid-range protocols show a significant mobility dependence on input load. Sage uses a 5 ug genomic sample with a broad size distribution for calibration. Higher input loads and samples with narrow size distributions will require user optimization of programmed size values for accurate results.
- Input loads higher than 10 ug per lane will have unpredictable results, and are not recommended.
If you plan to use the the BluePippin for mate-pair library construction, we’d love to hear of your progress or suggestions. To share, please contact us at info@sagescience.com or reply to this blog.
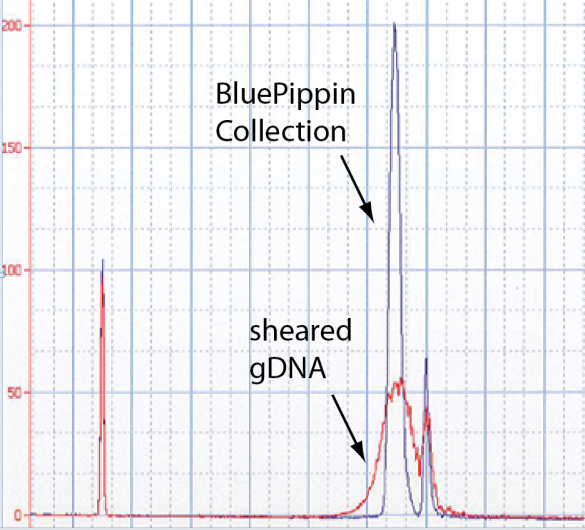
The image below is an Agilent Bioanalyzer trace of a nice 8 kb collection in the middle of a gDNA shear. This is near the limit of the Bioanalyzer’s detection target range.
Publication: Spike-In Standards for Managing Systematic Sequencing Errors
Here’s a paper worth checking out: “Synthetic Spike-in Standards Improve Run-Specific Systematic Error Analysis for DNA and RNA Sequencing” from lead author Justin Zook at the National Institute of Standards and Technology. Published in PLoS One last month, the paper describes a study of ways to better manage systematic errors in DNA or RNA sequencing.
Most sequencing work currently relies on algorithms that recalibrate base scores after calculating a correction factor using either a subset of the sequenced data set or a separate data set, the paper’s authors write. They propose using synthetic spike-in standards, in this demonstration using RNA spike-ins for sequencing human RNA. This is followed up with base recalibration with the Genome Analysis Toolkit (GATK from the Broad Institute) that more accurately adjusts based on the spike-in’s unique sequence signature.
“Compared to conventional GATK recalibration that uses reads mapped to the genome, spike-ins improve the accuracy of Illumina base quality scores by a mean of 5 Phred-scaled quality score units, and by as much as 13 units at CpG sites,” the authors write. “In addition, since the spike-in data used for recalibration are independent of the genome being sequenced, our method allows run-specific recalibration even for the many species without a comprehensive and accurate SNP database.”
In a paper that focuses on improving quality and uniformity, we were delighted to see that our Pippin platform was used for the cDNA size selection step with Illumina sequencing.
Congratulations to authors Justin Zook, Daniel Samarov, Jennifer McDaniel, Shurjo Sen, and Marc Salit!