A novel approach for enriching mitochondrial DNA for sequence analysis without PCR or gradient centrifugation.
View or download the full Application Note here.
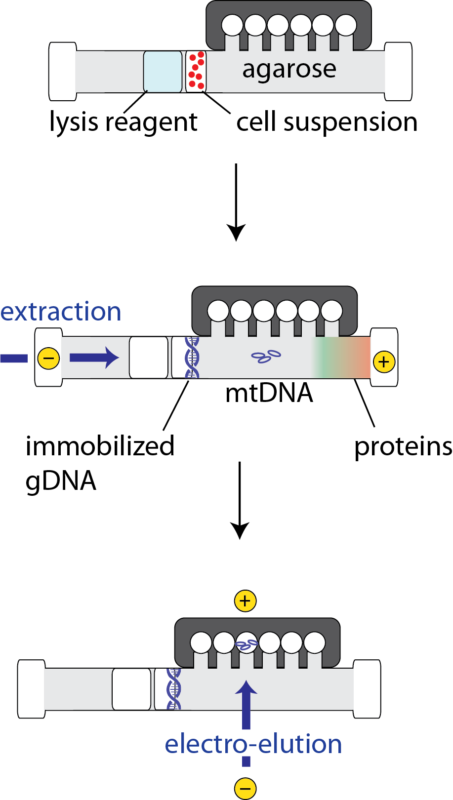
To ensure detection of low abundance mtDNA mutations without interference from nuclear pseudogenes, many researchers enrich mitochondria using differential gradient centrifugation, or enrich specific mitochondrial DNA sequences by PCR, before NGS sequence analysis. We demonstrate a novel approach to mtDNA isolation using the SageHLS platform which provides enrichment levels comparable to gradient centrifugation, but with a small fraction of the hands-on time.
A Schematic of mtDNA extraction process on the SageHLS gel cassette:
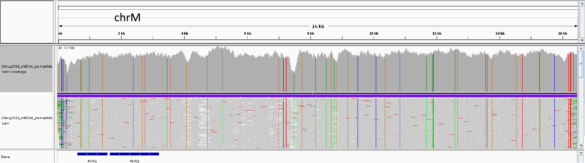
IGV view of sequence data from Illumina Miseq obtained from HLS-enriched mtDNA sample. The upper panel shows coverage and the lower panel shows line graphs for the individual sequencing reads. Coverage was approximately ~16,000-fold:
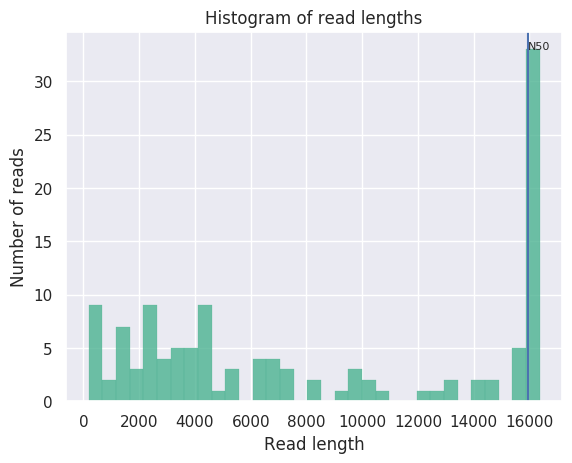
Length distribution of reads from Oxford Nanopore Minion sequencing of HLS-enriched mtDNA. Overall coverage was about 60-fold. The histogram shows that 30 of the reads were full-length 16.5kb reads of the entire mtDNA molecule:
Our results demonstrate that the SageHLS system can be used to enrich mtDNA from human cells to purity levels comparable to those offered by much more laborious “gold-standard” methods utilizing density gradient isolation of mitochondria prior to DNA isolation. Our method requires a suspension of 1-1.5m intact human (or other mammalian) cells. The concentration and purity of HLS-enriched mtDNA is compatible with mtDNA sequencing by Illumina platforms, and should enable detection of rare heteroplasmic mtDNA variants that are present at low single digit percentages. In addition, as input requirements for single molecule platforms decrease (Oxford Nanopore and PacBio), our method should be useful for preparing mtDNA inputs for those platforms, which will facilitate detection of larger indels and rearrangements.